Research
The Weber-Ban research group addresses the function, mechanism and substrate recruitment strategies of bacterial degradation complexes. Energy-dependent protein degradation ensures protein homeostasis in all organisms. It is carried out by bipartite molecular machines consisting of a central protease cylinder associating with ring-shaped regulator complexes responsible for substrate recruitment.
In mycobacteria, two types of cytosolic protein degradation systems exist, both under investigation in our group: the modular Clp protease complexes present in all bacteria, and a proteasome assembly unique to the actinobacterial phylum.
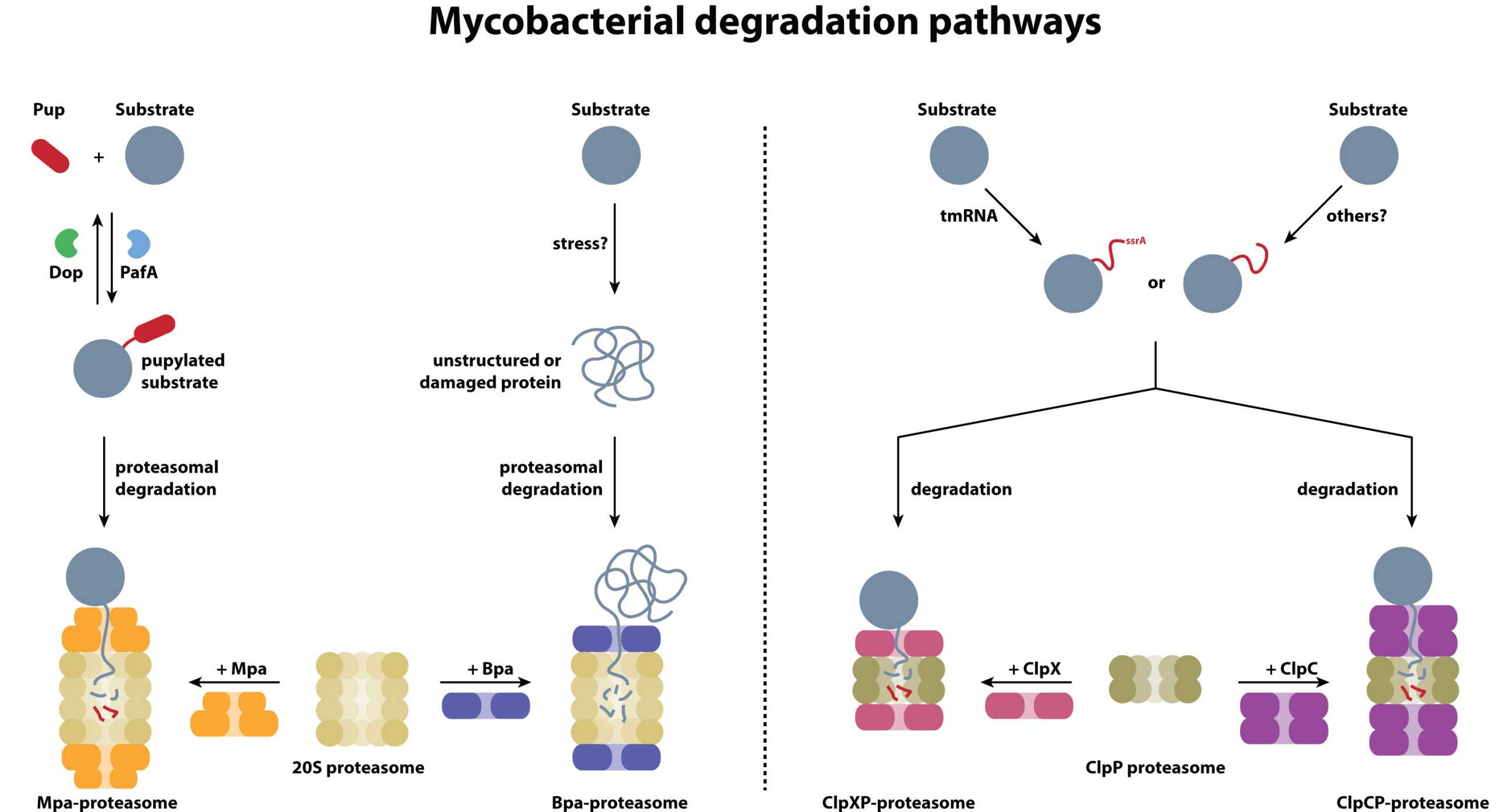
Substrates to the latter are recruited by a ubiquitin-like pathway termed pupylation, conjugating the small protein Pup (prokaryotic ubiquitin-like protein) to lysine residues of protein substrates.The proteolytic core particles of both protease systems can associate with different ring-shaped regulators.
ClpP interacts either with the ClpX chaperone ring featuring one ATPase module per protomer or with ClpC possessing two ATPase modules per subunit. Substrate profiles of ClpXP and ClpCP are different but partially overlapping. The proteasomal core particle can interact with the mycobacterial proteasomal ATPase Mpa (referred to as ARC in other actinobacteria) to degrade pupylated substrates or with the ATP-independent bacterial proteasome activator Bpa to recruit untagged but partially disordered substrates. Clp- and proteasome-mediated degradation have recently emerged as novel drug targets against Mycobacterium tuberculosis infections.